Base Boosting¶
Introduction¶
The standard formulation of gradient boosting begins forward stagewise additive modeling with a constant model, such as zero or the optimal constant model. Then, the loop body generates sequential improvements to the constant model. As such, the standard formulation of gradient boosting is an entirely data-driven method that derives a regressor exclusively from the statistical evidence present in the training examples itself.
In constrast, base boosting begins forward stagewise additive modeling with precomputed estimates from an existing, possibly nonconstant, model, which may contain prior knowledge or physical insight about the underlying data generating mechanism. Then, the loop body generates sequential improvements to the existing model. As such, this formulation of gradient boosting enables an existing model to precondition functional gradient descent, which is the underlying first-order optimization algorithm.
Example¶
Here, we consider an example that compares nested and non-nested cross-validation procedures in a quantum computing regression task. We plan to perform thirty random trials:
n_trials = 30
wherein each random trial uses base boosting. We begin by loading the training examples:
from physlearn.datasets import load_benchmark
# Shapes of (95, 5) and (41, 5), respectively.
X_train, _, y_train, _ = load_benchmark(return_split=True)
Next, we select a single basis function:
n_regressors = 1
basis_fn = 'stackingregressor'
stack = dict(regressors=['ridge', 'randomforestregressor'],
final_regressor='kneighborsregressor')
which is StackingRegressor with first layer regressors
Ridge and RandomForestRegressor and final layer regressor
KNeighborsRegressor from scikit-learn.
We select the squared error loss function for the pseduo-residual computation:
boosting_loss = 'ls'
and we select the line search options:
line_search_options = dict(init_guess=1, opt_method='minimize',
method='Nelder-Mead', tol=1e-7,
options={"maxiter": 10000},
niter=None, T=None, loss='lad',
regularization=0.1)
We bundle the options into a dict:
base_boosting_options = dict(n_regressors=n_regressors,
boosting_loss=boosting_loss,
line_search_options=line_search_options)
We choose the (hyper)parameters to exhaustively search over in the non-nested cross-valdation procedure and in the inner loop of the nested cross-validation procedure. Namely, the regularization strength in ridge regression, the number of decision trees in random forest, and the number of neighbors in k-nearest neighbors:
search_params = {'reg__0__alpha': [0.5, 1.0, 1.5],
'reg__1__n_estimators': [30, 50, 100],
'reg__final_estimator__n_neighbors': [2, 5, 10]}
Then, we make an instance of the regressor object using the aforespecified choices:
from physlearn import Regressor
reg = Regressor(regressor_choice=basis_fn, stacking_options=dict(layers=stack),
target_index=4, scoring='neg_mean_absolute_error',
base_boosting_options=base_boosting_options)
where the target index corresponds to the fifth single-target regression subtask.
We make arrays to store the nested and non-nested cross-validation scores:
import numpy as np
non_nested_scores = np.zeros(n_trials)
nested_scores = np.zeros(n_trials)
and we also import KFold from scikit-learn:
from sklearn.model_selection import KFold
We start the random trials and collect the scores:
for i in range(n_trials):
# Make two instances of k-fold cross-validation, whereby
# we generate the same indices for non-nested cross-validation
# and the outer loop of nested cross-validation.
outer_cv = KFold(n_splits=5, shuffle=True, random_state=i)
inner_cv = KFold(n_splits=5, shuffle=True, random_state=i)
# Perform a non-nested cross-validation procedure.
reg.search(X=X_train, y=y_train, search_params=search_params,
search_method='gridsearchcv', cv=outer_cv)
non_nested_scores[i] = reg.best_score_
# Perform a 5*5-fold nested cross-validation procedure.
outer_loop_scores = reg.nested_cross_validate(X=X_train, y=y_train,
search_params=search_params,
search_method='gridsearchcv',
outer_cv=outer_cv,
inner_cv=inner_cv,
return_inner_loop_score=False)
nested_scores[i] = outer_loop_scores.mean()
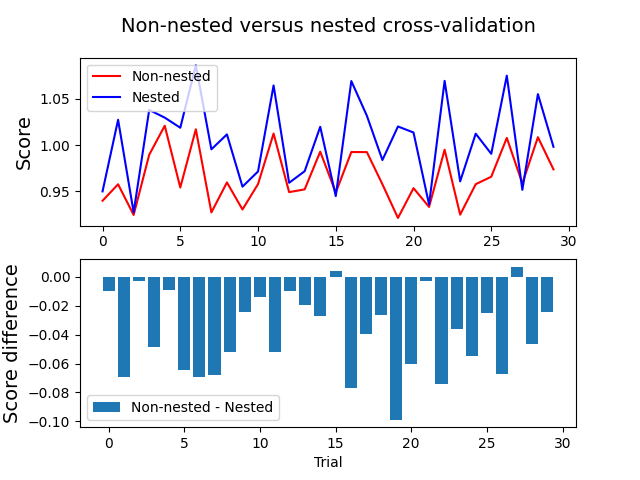
Lastly, we plot the nested and non-nested cross-validation scores, as well as the score difference, for each of the 30 random trials:
from physlearn.supervised import plot_cv_comparison
plot_cv_comparison(non_nested_scores=non_nested_scores, nested_scores=nested_scores,
n_trials=n_trials)
which outputs:
Average difference of -0.038677 with standard deviation of 0.027483.
References¶
Alex Wozniakowski, Jayne Thompson, Mile Gu, and Felix C. Binder. “A new formulation of gradient boosting”, Machine Learning: Science and Technology, 2 045022 (2021).
John Tukey. “Exploratory Data Analysis”, Addison-Wesley (1977).
Jerome Friedman. “Greedy function approximation: A gradient boosting machine,” Annals of Statistics, 29(5):1189–1232 (2001).
Trevor Hastie, Robert Tibshirani, and Jerome Friedman. “The Elements of Statistical Learning”, Springer (2009).